graph LR;
A(Human Aligned<br>BAM) --> |Filter and convert| B(Non Human Reads<br>FASTQ)
B --> |Trimmomatic| C(Clean FASTQ File)
C --> |MetaPhlAn| D(MetaPhlAn 4 /<br>vJan21 Abundances<br>Tabular)
C --> |HumanN| E(HumanN 3.6 genes abundance)
C --> |HumanN| F(HumanN 3.6 Pathways abundance)
C --> |HumanN| G(HumanN 3.6 Pathways coverage)
C --> |FastQC| H(Reads QC report - after)
B ---> |FastQC| I(Reads QC report - before)
070-oral_microbiome
Oral microbiome dataset
Description
The collection of buccal swabs for human genetic sequencing presents an advantageous opportunity to simultaneously explore the oral microbiome using the non-human DNA sequences found. This metagenomic analysis provides insight into the diverse microbial community within the buccal mucosa. The obtained data not only enriches our genetic understanding but also broadens our knowledge on the microorganisms residing in the oral cavity, their interactions, and potential implications for oral and systemic health. This dual analysis underscores the utility of buccal swabs as a resourceful means for both genetic and microbial investigations.
Introduction
The primary intent behind collecting buccal swabs was to explore human genetics. Concurrently, it yielded metagenomic data, unveiling the microbial community within the oral cavity.
This data illuminates the diversity and interactions of microorganisms, specifically in the buccal mucosa. Delving into the oral microbiome presents an opportunity to understand its linkage to health conditions. For instance, an imbalanced oral microbiome can lead to periodontal disease, associated with an increased risk of cardiovascular diseases. Moreover, alterations in the oral microbiome have been observed in oral cancers, hinting at its potential role in their onset and progression The oral microbiome: diversity, biogeography and human health.
Through this data, the exploration into possible connections between oral microbial composition and various health conditions is facilitated, marking a step towards understanding the broader health implications.
Measurement protocol
This dataset is a derivative of the human genome dataset (that has been collected via buccal swabs) - ergo the measurement protocol can be found at the Human genetics dataset.
Data availability
The information is stored in multiple parquet files:
oral_microbiome.parquet: Sequencing and QC statistics.metaphlan_*: 8 tables with MetaPhlAn 4 vJan21 relative abundances, separated by taxonomic levels.humann_aggregated_*: arrow files of either gene level abundances or pathway level abundances+coverage from HumanN 3.6 given MetaPhlAn 4 vJan21 outputs
Relevant links
from pheno_utils import PhenoLoaderpl = PhenoLoader('oral_microbiome')
plPhenoLoader for oral_microbiome with
39 fields
2 tables: ['oral_microbiome', 'age_sex']Data dictionary
pl.dict| folder_id | feature_set | field_type | field_string | description_string | relative_location | data_coding | stability | units | sampling_rate | bulk_file_extension | bulk_dictionary | strata | sexed | array | debut | completed | transformation | list_of_tags | pandas_dtype | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tabular_field_name | ||||||||||||||||||||
| collection_date | NaN | oral_microbiome | Datetime | Sampled date | Date sample was given | oral_microbiome/oral_microbiome.parquet | NaN | Complete | Time | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | datetime64[ns] |
| metaphlan_abundance_kingdom_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (kingdom) | Aggregated abundance (MetaPhlAn 4) at the king... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_phylum_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (phylum) | Aggregated abundance (MetaPhlAn 4) at the phyl... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_class_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (class) | Aggregated abundance (MetaPhlAn 4) at the clas... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_order_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (order) | Aggregated abundance (MetaPhlAn 4) at the orde... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_family_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (famiily) | Aggregated abundance (MetaPhlAn 4) at the fami... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_genus_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (genus) | Aggregated abundance (MetaPhlAn 4) at the genu... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_species_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (species) | Aggregated abundance (MetaPhlAn 4) at the spec... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan_abundance_strain_parquet | NaN | metaphlan_aggregated | Mapping table (group) | MetaPhlAn 4 abundance (strain) | Aggregated abundance (MetaPhlAn 4) at the stra... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| raw_fastq | NaN | raw_fastq | Fastq file (individual) | Raw FASTQ path (Post human removal) | The original FASTQ file | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | fastq.gz | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| trimmed_fastq | NaN | trimmed_fastq | Fastq file (individual) | Post QC FASTQ path | FASTQ file post Trimmomatic (removing adaptors... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | fastq.gz | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| fastqc_raw_html | NaN | fastqc | Bulk file (individual) | FASTQC HTML path Raw data | FASTQC Output for Raw FASTQ - HTML | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | html | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| fastqc_raw_zip | NaN | fastqc | Bulk file (individual) | FASTQC zip path Raw data | FASTQC Output for Raw FASTQ - Zip | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | zip | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| fastqc_trimmed_html | NaN | fastqc | Bulk file (individual) | FASTQC HTML path Trimmed data | FASTQC Output for Post QC FASTQ - HTML | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | html | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| fastqc_trimmed_zip | NaN | fastqc | Bulk file (individual) | FASTQC zip path Trimmed data | FASTQC Output for Post QC FASTQ - Zip | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | zip | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| metaphlan4_results_tsv | NaN | metaphlan | Bulk file (individual) | MetaPhlAn 4 abundance path | MetaPhlAn4 relative abundances | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | tsv | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| raw_read_count | NaN | oral_microbiome | Integer | Raw reads count | # Reads in the original bam file from gencove | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | int |
| non_human_read_count | NaN | oral_microbiome | Integer | Non-human reads count | # Reads with no alignment to the human genome | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | int |
| trimmed_non_human_read_count | NaN | oral_microbiome | Integer | Post QC Non-human reads count | # Non human Reads After QC | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | int |
| metaphlan4_aligned_read_count | NaN | oral_microbiome | Integer | Jan21 MetaPhlAn4 unique aligned reads count | # Reads after QC aligned to bacteria genomes | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | int |
| gene_aligned_read_count | NaN | oral_microbiome | Integer | HumanN 3.6 diamond aligned unique read count | # Reads after QC aligned to Uniref90 | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | NaN | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | int |
| humann_gene_families_tsv | NaN | humann | Bulk file (individual) | HumanN 3.6 - Gene families in original .tsv | HumanN 3.6 output of Microbial reads protein a... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | tsv | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_gene_familites_gene_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Gene families, in gene only level... | HumanN 3.6 output of Microbial reads protein a... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_gene_familites_microbe_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Gene families, microbiome level, ... | HumanN 3.6 + MetaPhlAn 4 output of Microbial r... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_abundance_tsv | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway abundance in original .tsv | HumanN 3.6 output of Oral microbime metaboliti... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | tsv | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_abundance_pathway_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway abundance, in gene only l... | HumanN 3.6 output of Oral microbime metaboliti... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_abundance_microbe_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway abundance, microbiome lev... | HumanN 3.6 + MetaPhlAn 4 output of Oral microb... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_coverage_tsv | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway coverage in original .tsv | HumanN 3.6 output of Oral microbime metaboliti... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | tsv | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_coverage_pathway_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway coverage, in gene only le... | HumanN 3.6 output of Oral microbime metaboliti... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_pathway_coverage_microbe_level_parquet | NaN | humann | Bulk file (individual) | HumanN 3.6 - Pathway coverage, microbiome leve... | HumanN 3.6 + MetaPhlAn 4 output of Oral microb... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | parquet | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_intermediate_files_tar | NaN | humann | Bulk file (individual) | HumanN 3.6 - Intermidiate files | All of the HumanN 3.6 + MetaPhlAn 4 intermidia... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | tar.gz | NaN | Supporting | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_gene_familites_gene_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Gene families (gene le... | Aggregated HumanN 3.6 output of Microbial read... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_gene_familites_microbiome_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Gene families (microbe... | Aggregated HumanN 3.6 output of Microbial read... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_pathway_abundance_pathway_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Pathways abundances (p... | Aggregated HumanN 3.6 output of Oral microbime... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_pathway_abundance_microbiome_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Pathways abundances (m... | Aggregated HumanN 3.6 output of Oral microbime... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_pathway_coverage_pathway_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Pathways coverage (pat... | Aggregated HumanN 3.6 output of Oral microbime... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
| humann_aggregated_pathway_coverage_microbiome_level_arrow | NaN | humann_aggregated | Mapping table (group) | Aggregated HumanN 3.6 - Pathways coverage (mic... | Aggregated HumanN 3.6 output of Oral microbime... | oral_microbiome/oral_microbiome.parquet | NaN | Complete | NaN | NaN | arrow | NaN | Primary | Both sexes | Single | NaN | NaN | NaN | Oral Microbiome | string |
Plot histogram and ecdf for raw_read_count at baseline visit
from pheno_utils.basic_plots import hist_ecdf_plots
col = "raw_read_count"
df = pl[[col] + ["age", "sex", "collection_date"]].loc[:,:,"00_00_visit",0,:]
# plot histogram and ecdf
hist_ecdf_plots(df.dropna(subset=[col,"sex", "age"]), col, gender_col="sex")
# stats
display(df[col].describe().to_frame().T)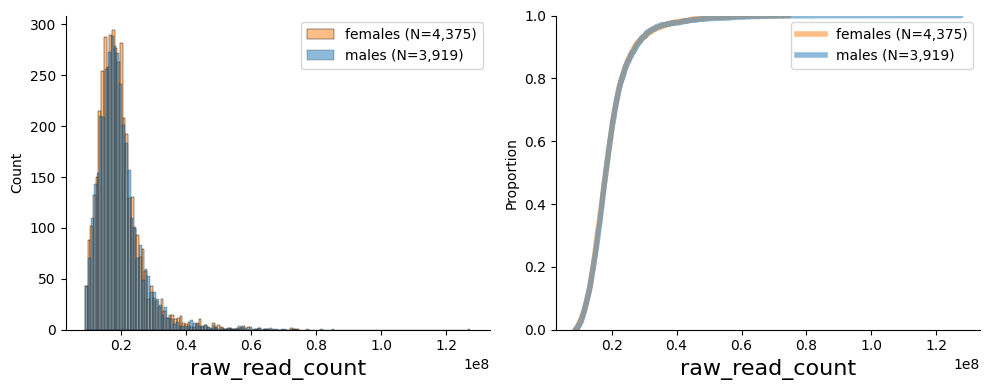
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| raw_read_count | 8294.0 | 19529470.245479 | 7338498.393217 | 8805038.0 | 15039265.5 | 18221025.5 | 21923790.0 | 127497332.0 |